Announcing {plumber2mcp} and {hgnc.mcp}: of model context protocol and vibe coding
On Nov 4, 2025, I received an e-mail from Claude letting me know that I’d been granted $1,000 in credits for the web-based Claude Code environment (https://claude.ai/code). The credits were originally set to expire on Nov 18, 2025 (later extended to Nov 23, 2025):
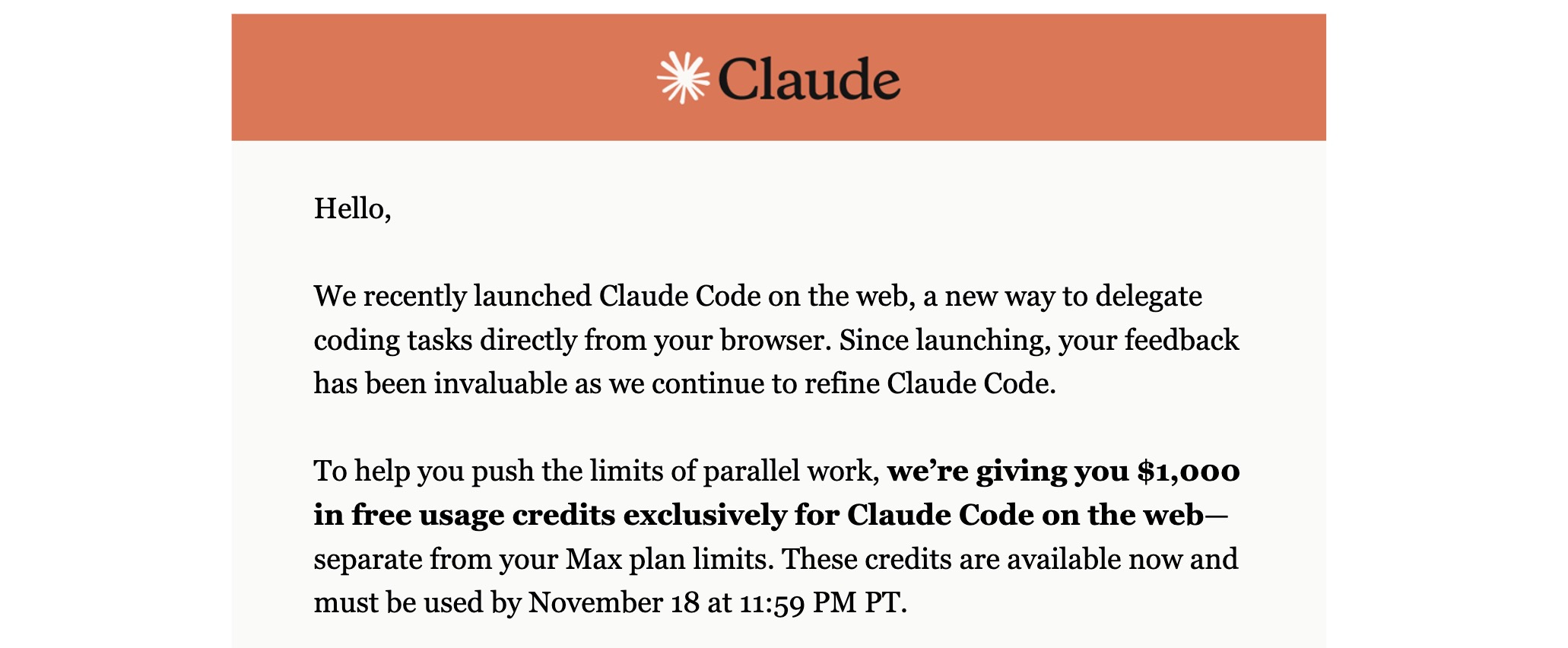
Naturally, I decided to make the most of this rare opportunity. I pulled out a few coding projects that had been sitting on the backburner and started vibe-coding as intensely as I could. Surprisingly, despite furiously coding between Nov 4 and Nov 18, I only managed to burn through $89 of the $1,000. I was convinced that such heavy usage would drain the credits quickly, but apparently not:

{plumber2mcp}: An R package to help convert a plumber API into an MCP server
One of my first experiments was inspired by FastAPI-MCP. I loved the idea of taking an existing FastAPI application and essentially “turning it into” an MCP server. I wanted to do the same for R: take a plumber API and expose its endpoints as MCP utilities. It also seemed like a good excuse to get some hands-on exposure to MCP, because at the time, the protocol felt confusing and abstract.
I’m a long-time fan of plumber and have used it extensively to build internally facing APIs. So back in July, when I first got access to Claude Code, I started with a simple prompt: “Build an R package that takes a plumber object and exposes its endpoints via MCP.” To make things easier for Claude—since R isn’t its strongest suit—I prepared the package skeleton with usethis.
Within a few days of intermittent vibing, I had a minimally functioning package. Testing the MCP side was more painful than expected, especially the requirement of wrapping HTTP endpoints with a script to support STDIO transport mode. It wasn’t shocking given how early MCP tooling still is, but I did hope for something a bit more plug-and-play. Hopefully the package documentation will make the experience smoother for others who want to build MCP tools.
Here’s what a typical plumber → MCP conversion looks like using plumber2mcp:
library(plumber)
library(plumber2mcp)
# Read API endpoint definitions, add MCP endpoints, and then run it
pr("plumber.R") %>%
pr_mcp(transport = "http") %>%
pr_run(port = 8000)
That first project taught me a lot about MCP. It was fun to watch trivial functions show up inside the MCP inspector and behave as expected. But I knew that for a serious application, I’d eventually need to revisit and refine the package.
{hgnc.mcp}: A combined API and MCP server to ease integration of HGNC resources with LLMs
As a bioinformatics researcher, I rely heavily on the HUGO Gene Nomenclature Committee (HGNC). LLMs—ChatGPT, Claude, and others—are generally knowledgeable about human genes, but they often struggle with alias resolution, gene families, or external IDs. This becomes a real problem during deep research tasks, where a model can get stuck on an outdated synonym or miss information due to a recent name change.
Two years ago, I attempted to address this by building a custom GPT called Hugo - Bioinformatics helper. It worked well for normalization and lookup tasks, but it was locked inside ChatGPT and couldn’t be integrated into other LLMs.
Just got GPTs enabled for my account and, of course, the first GPT I tried to build is an @genenames-savvy assistant.
— B. Arman Aksoy (@armish) November 9, 2023
Super excited to finally have a companion who can help me with one-off mundane ID mapping/look-up tasks pic.twitter.com/LZYb9ffeGT
With the free Claude credits acting as a catalyst, I wanted to revisit the idea—this time as an MCP server. I created an empty repository, opened a conversation with Claude, and outlined the plan:
- build an R package depending on
plumber2mcp, - expose HGNC resources as MCP tools,
- implement alias mapping, external ID lookups, gene family retrieval, etc.
I asked Claude not to write code immediately but instead to design the plan as a TODO.md. For each item in that TODO list, I opened a fresh conversation, let it generate a PR, and merged when things looked good.
Working on an R project inside Claude Code Web has limitations. The environment includes basic Python but no R installation, which means most of the R code was produced completely “blind.” Actual testing mostly happened in GitHub Actions, so I ended up bouncing between action logs and Claude Code Web—definitely not the smoothest workflow. On the command line, Claude Code can leverage your local environment and iterate much faster.
But still, I was impressed with how much blind coding actually worked. Most tests passed on the first try, and when something failed, pasting the error into Claude was usually enough to fix it on the spot.
This wasn’t a small project either. It required:
- creating a proper R package,
- learning how to work with HGNC’s TSV files,
- providing a suite of functions,
- decorating them with
plumberannotations, - exposing them as MCP tools/resources,
- and testing everything end-to-end.
Ten days of intense vibe-coding later, I had spent only $81 and ended up with a solid R package that did exactly what I needed.

When it was time to test integration with LLMs, things got complicated again. I wanted to use Claude Desktop, but expecting normal users to install R just to access HGNC data wasn’t realistic. So I decided to ship everything as a Docker image and use the STDIO transport. This required switching to Claude Code CLI because waiting for GitHub Actions to build and push images before locally testing was too slow.
Once the Docker workflow was set up, I discovered that my earlier plumber2mcp implementation was based on an outdated MCP specification. Updating it to the newer version wasn’t too bad, though, and before long, I had everything running smoothly in Claude Desktop:
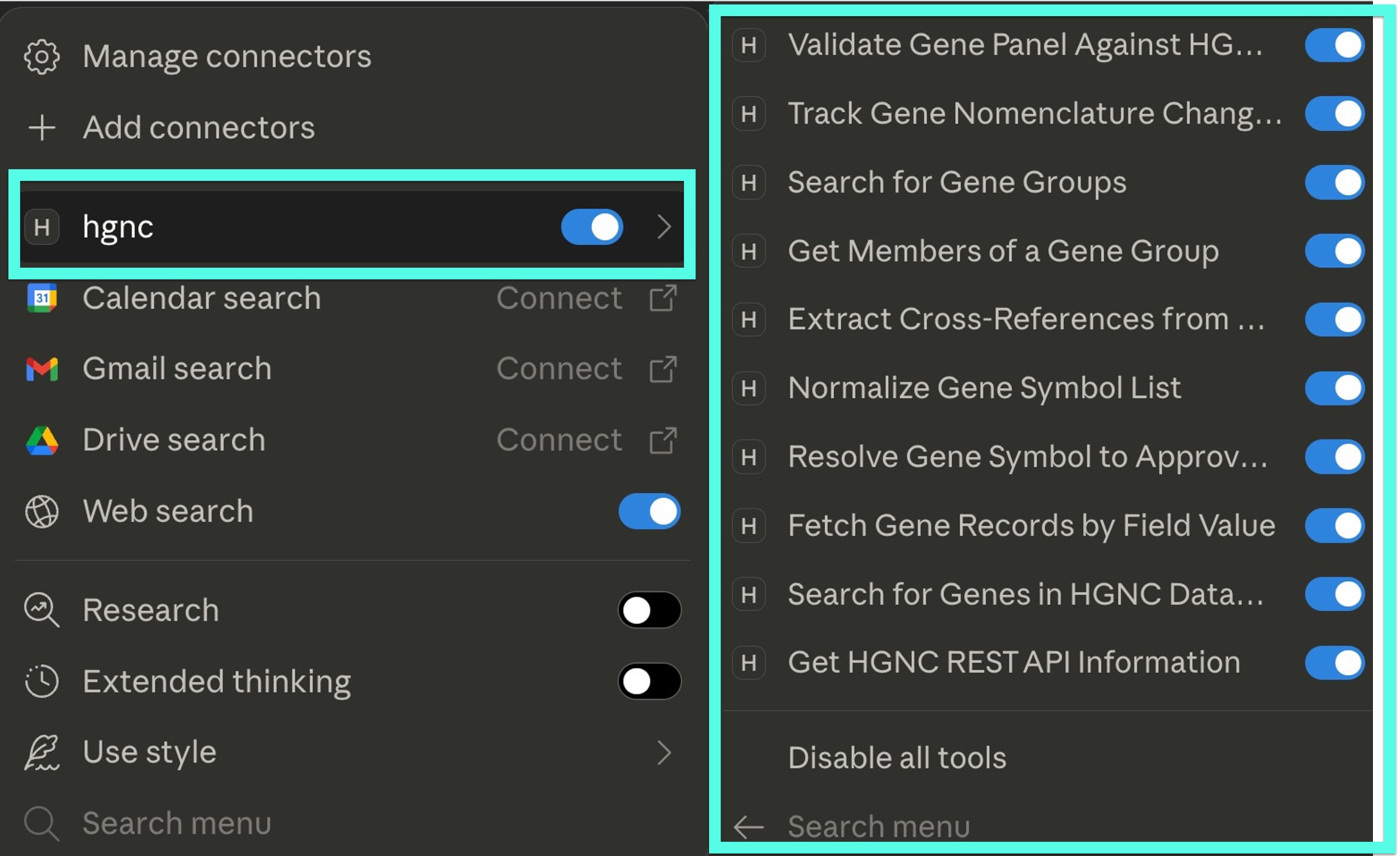
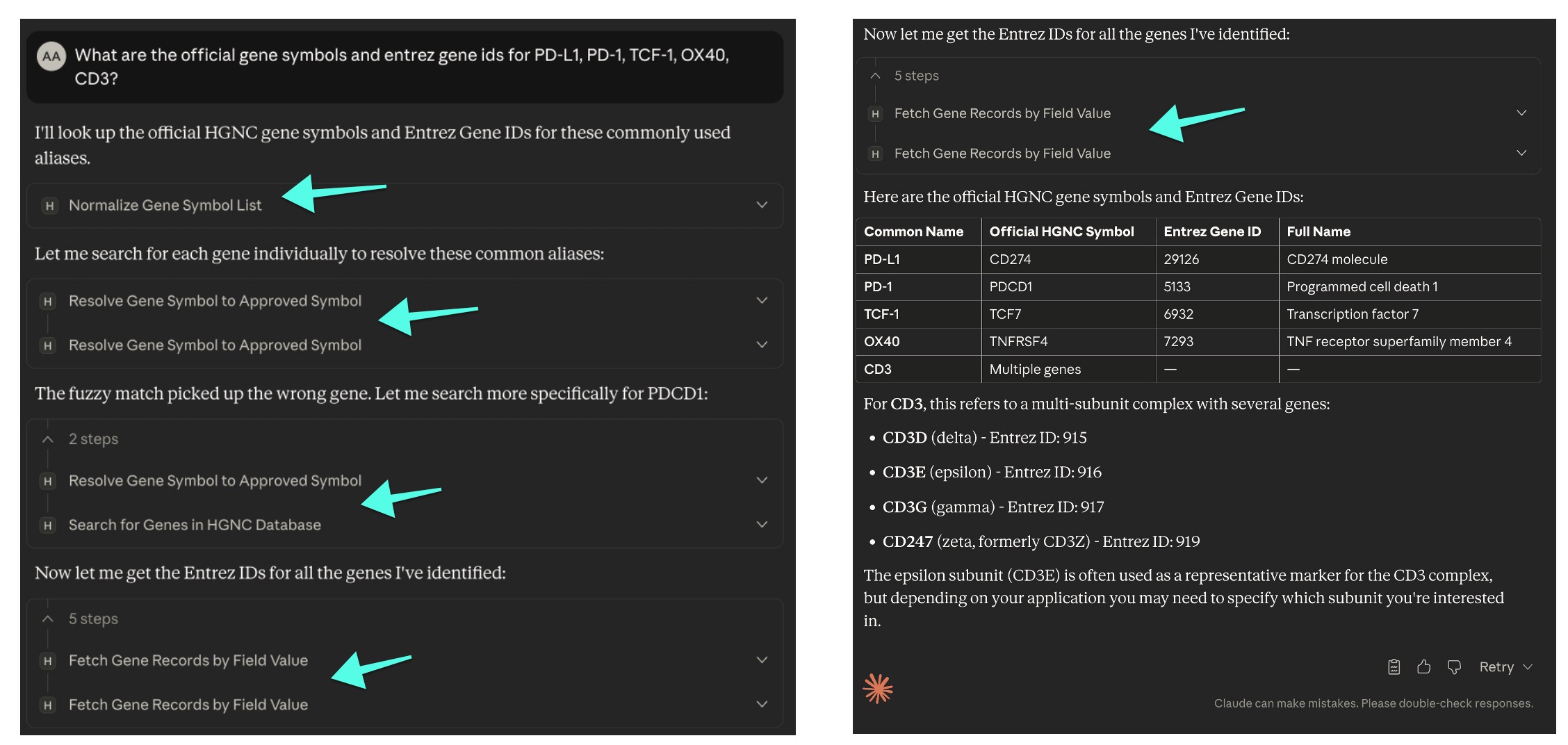
Here’s an example showing how the HGNC MCP server resolves a mix of aliases without needing explicit instructions—and without worrying about outdated knowledgebases:
What are the official gene symbols and entrez gene ids for PD-L1, PD-1, TCF-1, OX40, CD3?
If you’re a Claude Desktop user and want to try it yourself, first pull the latest Docker image:
$ docker pull ghcr.io/armish/hgnc.mcp:latest
Then update your Claude configuration (on macOS this lives at ~/Library/Application\ Support/Claude/claude_desktop_config.json):
{
"mcpServers": {
"hgnc": {
"command": "docker",
"args": ["run", "--rm", "-i", "-v", "hgnc-cache:/home/hgnc/.cache/hgnc", "ghcr.io/armish/hgnc.mcp:latest", "--stdio"]
}
}
}
After restarting Claude Desktop, you should see hgnc under your tools menu, where you can toggle individual tools on or off.
Final thoughts
What did I learn from this extended vibe-coding adventure?
- It has never been this easy to go from a rough idea to a functional solution. If the credits were coming out of my own pocket, it might feel different—but for ~$90, the experience was absolutely worth it.
- Building an MCP solution is not trivial. It spans multiple layers (R package → API → MCP → STDIO wrapping → Dockerization → LLM configuration) and requires constant context-switching.
- Getting an MCP server to “work” is only half the story: a couple of queries can blow through the conversation’s context window. Optimizing MCP responses for token efficiency is its own challenge.
- Claude Code Web is great for lightweight work, but blind-coding R packages gets clumsy fast. Claude Code CLI is significantly smoother because it can run things locally and iterate quickly. If only my free credits applied there too.
- The limbo periods while LLMs are thinking are real. For quick tasks, the pauses are manageable. But during long vibe-coding sessions, the waiting becomes mentally draining. Session blocks help, but it’s still easy to burn out.
- Publishing an R package on CRAN requires patience. I submitted an early version of
plumber2mcpalmost two weeks ago and still haven’t heard back. Until then, both packages are available on GitHub and can be installed viadevtools,remotes, etc.